Home > Database
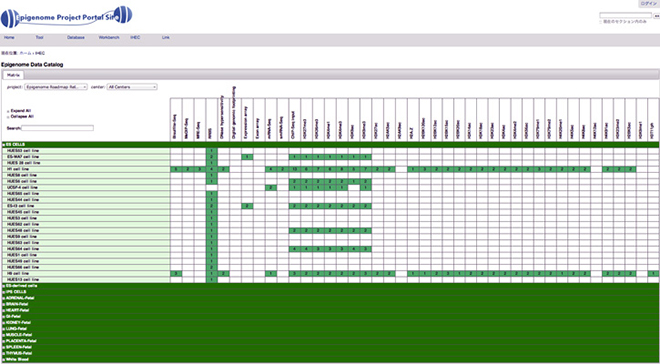
 Database
Database
 Feature
Feature
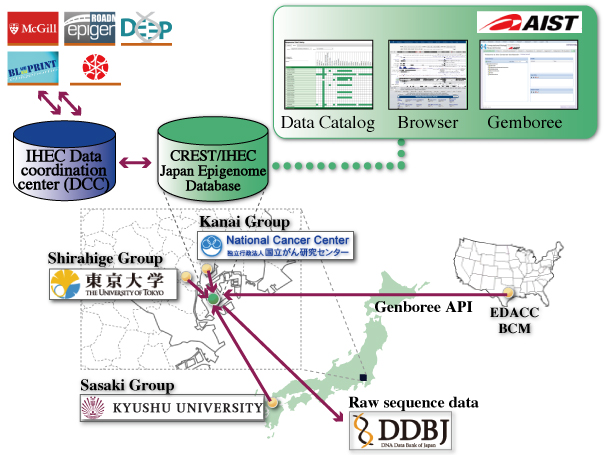
- The database will have a flexible data process and analytical pipelines specialized for Japanese IHEC members.
- Raw sequence data will be registered at the DNA Data Bank of Japan, DDBJ; DNA Data Bank of Japan.
*The database will be publicly accessible and contain only data that cannot be aggregated to generate a dataset unique to an individual. - Suzuki A, Kawano S, Mituyama T, Suyama M, Kanai Y, Shirahige K, Sasaki H, Tokunaga K, Tsuchihara K, Sugano S, Nakai K, Suzuki Y.
DBTSS/DBKERO for integrated analysis of transcriptional regulation.
Nucleic Acids Res. (2018) 46, D229–D238.
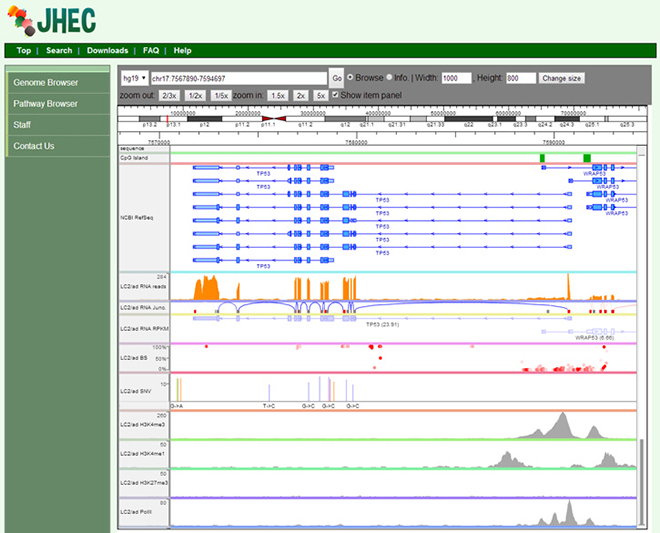
 Browser
Browser
 Organization
Organization
- Developers of our Database:
Toutai Mituyama, National Institute of Advanced Industrial Science and Technology (AIST)
Yutaka Suzuki, University of Tokyo
Mikita Suyama, Kyushu University
*Toutai Mituyama and Yutaka Suzuki are members of the Data Ecosystem Working group, IHEC - Producers of Data: Team Kanai、Team Shirahige、and Team Sasaki
- Bioethics Guideline: Yae Kanai, National Cancer Center Research Institute
*Yae Kanai is a member of the Bioethics Workgroup, IHEC